Abstract
CEBPA is a bZIP transcription factor mutated in approximately 10% of sporadic acute myeloid leukaemia (AML). In 50% of cases a single allele of CEBPA is mutated while in the other 50% both alleles are mutated. The majority of dm-CEBPA mutations occur with one allele mutated at the N terminal region (CEBPANterm) resulting in a frame-shift, and the other allele at the C terminus (CEBPACterm), leading to inframe indels in the bZIP/Leucine zipper region of the protein, presumably abrogating DNA binding. In wild-type conditions two CEBPA isoforms are translated; full length p42 in far greater amounts than p30, an N-terminal truncated isoform derived from an in frame internal initiating site.
In addition to somatic mutations of CEBPA in AML, several kindreds have been described who carry germline mutations in CEBPANterm. This results in familial predisposition to AML. Such cases highlight a strong selective pressure to acquire CEBPA CEBPACterm, as not only do the vast majority of patients acquire such mutations at presentation with AML, but several have shown distinct CEBPACterm mutations at relapse. CEBPACterm are often considered null with respect to DNA binding capacity, implying that CEBPA functional activity as a transcription factor arises from its p30 isoform in cases with mutations in both alleles.
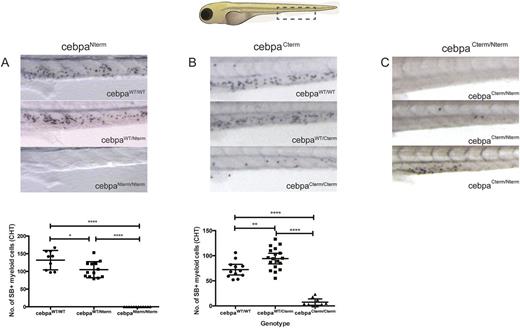
To model the steps leading to leukemia development in CEBPA-mutated AML we used TALENs to generate zebrafish carrying analogous CEBPANterm (K75fs; cebpaNterm) and CEBPACterm (K243del243-246, cebpaCterm). To assess the affect of these mutation (alone or in combination) on developmental myelopoiesis we used Sudan Black (SB) staining at 5 days post fertilization (dpf). cebpaNterm/Nterm mutants, showed a complete absence of SB+ cells by 5dpf, while cebpaNterm/+ mutants had modestly, but statistically significantly reduced numbers (A). To further delineate the stage in myeloid differentiation that the observed defect arose we used flow cytometry of cebpaNterm mutants carrying the Tg(Lys:mCherry) and Tg(pu.1:GFP) transgenes . At 5 dpf we observed that both pu.1 (early myeloid) and lysC (later myeloid) expressing cells could be seen in all genotypes. Nonetheless, we observed that the earliest myeloid cells (pu.1-GFP+/lysC mCherry-) and intermediate differentiation stage populations (pu.1-GFP+/lysC mCherry+) both showed dose dependent reduction in numbers with acquisition of cebpaNterm mutation. This data suggests that cebpaNterm mutation leads to a defect in myeloid differentiation in a allele dependent manner.
cebpaCterm mutants notably showed a distinct phenotype. Whilst cebpaCterm/Cterm showed severely reduced numbers of SB+ cells, in contrast to cebpaNterm/Nterm they were not completely absent (B). Furthermore, the cebpaCterm/+ mutants showed a significant increase in SB+ cells, in striking contrast to the reduction in cebpaNterm/+ (B). This suggests secondary CEBPACterm could convey a proliferation advantage to overcome the deficit in mature myeloid numbers induced by an initiating CEBPANterm. Analysis of cebpaCterm/+ and cebpa Nterm/+ mutantscarrying Tg(Lys:mCherry) show that the proportion of myeloid cells expressing the brightest mCherry are markedly reduced in cebpaCterm/+ mutants alone compared to WT siblings. Further, SB stains in cebpaCterm/+ and cebpaCterm/Cterm also show qualitative less intense and smaller granules suggesting defective granule formation in these mutants.
We then went on to determine the effects of combined mutation of cebpaCterm and cebpaNterm, since this is the most commonly observed genotype in patients. SB staining at 5dpf in cebpaNterm/Cterm showed a spectrum of phenotypes (C). These range from absent (upper) or severely reduced (middle) SB+ cells as in cebpaNterm or cebpaCterm respectively, or a of near normal numbers but with abnormal localization in the caudal haematopoietic tissue (fetal liver equivalent) (lower).
In conclusion we have generated zebrafish that model Cebpa N and C terminal mutations observed in AML and demonstrate differential and combined effects on developmental haematopoeisis arising from these 2 alleles. Our data suggest that mechanisms to overcome the myeloid differentiation defect resulting from CebpaNterm/+ may underpin the selective pressure driving mutations in the C term of the other Cebpa allele, which phenotypically compensate for this.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal